
 Web Content Display
Web Content Display
SOLARIS centre
 Web Content Display
Web Content Display
 Web Content Display
Web Content Display
Structural basis of transposon end recognition explains central features of Tn7 transposition systems
Transposases are proteins crucial for the mobility of transposons – genetic elements that can move within genomes. In prokaryotes, this process can lead to the horizontal transfer of virulence genes, contributing to the emergence of novel superbugs. SOLARIS Centre users have solved a 2.7 Å cryo-EM structure of prototypic Tn7 transposase TnsB interacting with the transposon end DNA.
The solved structure shows that TnsB binds DNA with sequence preference rather than strict sequence specificity, and that the formation of an array of TnsB molecules converts this preference into specific end recognition.
Cryo-EM structures
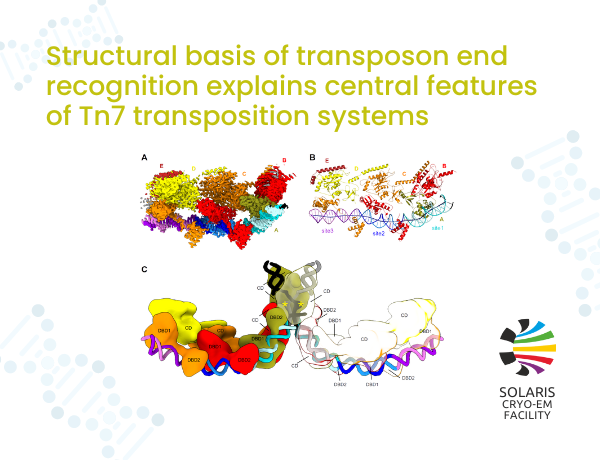
Scientists used cryoelectron microscopy (cryo-EM) to analyze complexes of a prototypic E. coli Tn7 TnsB transposase with double-stranded DNA substrates. For one of the substrates, which comprised three binding sites corresponding to the right end of the transposon, we observed the formation of long filaments that we used to solve the cryo-EM structure of the TnsB-DNA complex (Figure 1A, B).
In the structure, TnsB molecules interact with repeating binding sites in an intertwined fashion and adopt a beads-on-a-string architecture. The DNA-binding domains form only few base-specific contacts, leading to a DNA sequence preference rather than strict sequence specificity. The formation of an array of TnsB molecules, regulated by the overlap of TnsB-binding sites in the right end of the Tn7 element, converts this preference into specific end recognition. Users from International Institute of Molecular and Cell Biology in Warsaw proposed a model of the strand-transfer complex in which the terminal TnsB molecule is rearranged, so that its catalytic domain is in a position conducive to transposition (Figure 1C).
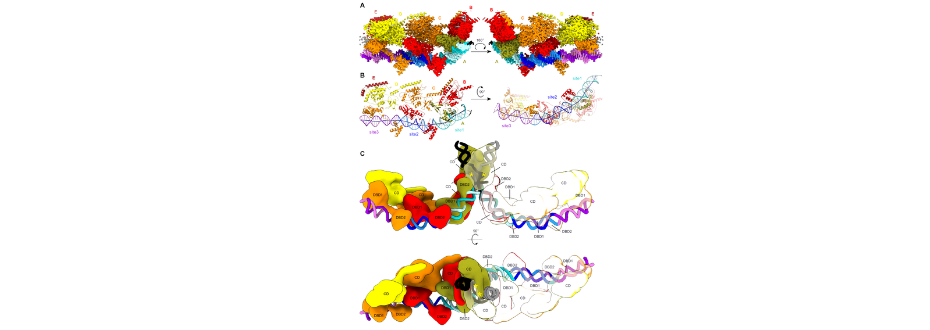
Figure 1. Structure of the TnsB-DNA complex. (A) Cryo-EM reconstruction of TnsB in complex with transposon end DNA. (B) Atomic model of TnsB bound to DNA. (C) Model of TnsB strand-transfer complex.
Next-generation gene editing tools
Some recently described transposons from the wide Tn7 family contain element-encoded CRISPR-Cas systems mediating RNA-guided transposition. They provide some of the most promising systems for next-generation gene editing tools and all encode TnsB-like transposases. The structural and biochemical analyses of Tn7 TnsB transposase and proposed model of the TnsB strand-transfer complex provide a mechanistic understanding of how TnsB molecules can interpret subtle differences in the spacing of diverged binding sites and explain central features of Tn7 transposition systems.
The publication available here:
Written by: Mariusz Czarnocki-Cieciura
