Web Content Display
Web Content Display
SOLARIS centre
 Web Content Display
Web Content Display
 Web Content Display
Web Content Display
Mechanism of a key protein complex in bacterial homologous recombination.
Scientists from the Laboratory of Protein Structure at the International Institute of Molecular and Cell Biology in Warsaw explained the mechanism of a key protein complex in bacterial homologous recombination.
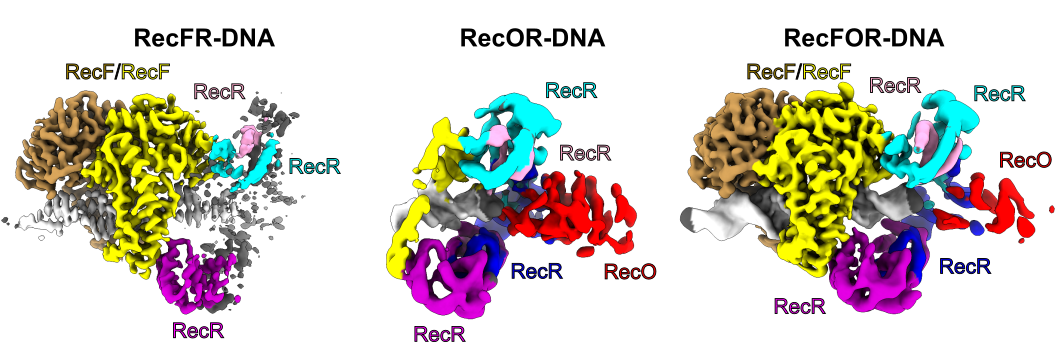
Abstract: Homologous recombination is one of the fundamental pathways for the repair of damaged genetic information. It involves the generation of single-stranded DNA at the site of damage. This DNA performs a homology-based search for the template. In bacteria, one of the homologous recombination-based repair pathways involves RecF, RecO and RecR proteins. They bind at the junction of single-stranded (ss) and double-stranded (ds) DNA and then facilitate the replacement of the SSB protein, which initially covers ssDNA, with RecA which promotes the search for the homologous sequence. However, the molecular mechanism of RecFOR cooperation in the pathway was largely unknown. Shivlee Nirwal and co-workers from Prof. Marcin Nowotny’s laboratory used the SOLARIS National Synchrotron Radiation Centre to determine the cryo-electron microscopy structures of the RecF-DNA and RecFOR-DNA complexes, and their subcomplexes, to elucidate the mechanism of the cooperation of these proteins in ss-dsDNA junction recognition.
Development: The RecF-DNA and RecFR-DNA subcomplex of the RecFOR-DNA assembly, show how the RecF dimer uses helical protrusions in its structure to bind double-stranded DNA. The RecFR-DNA subcomplex further shows the way one RecF protomer interacts with two different regions of a ring made of four RecR molecules which stabilize this ring on DNA. The lower-resolution reconstructions of the RecR–RecO subcomplex and the RecFOR–DNA assembly explain how RecO is positioned to interact with ssDNA and SSB, which is proposed to lock the complex on a ssDNA–dsDNA junction. This mode of RecFOR action is further supported by biochemical and biophysical experiments. Overall, the results obtained integrate and explain the wealth of biochemical data available for the RecFOR system and provide a framework for a complete understanding of the RecFOR homologous recombination pathway.
Figure 1. The cryo-EM reconstructions for the RecFOR-DNA assembly and the RecFR-DNA and RecOR-DNA subcomplexes of the assembly.
Written by: Shivlee Nirwal
The publication can be found here: