
 Web Content Display
Web Content Display
SOLARIS centre
 Web Content Display
Web Content Display
 Web Content Display
Web Content Display
Machine learning-based IR imaging model for pancreatic tissue recognition developed!
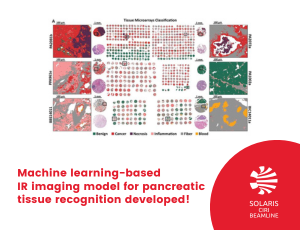
Researchers from CIRI beamline built the first IR-based machine learning model for comprehensive pancreatic histopathology, based on 600 needle biopsies from 250 patients. Total number of spectra collected with FT-IR imaging was equal to 672 407 632 giving one of the biggest dataset in tissue recognition collected up to this time.
Infrared (IR) imaging in combination with machine learning is a great tool for different cancers recognition. However, it was never used for pancreatic cancer recognition. Diagnosis of pancreatic cancer – because of the lack of characteristic disease symptoms - in the early stage of the disease is difficult. The gold standard in diagnosis is histopathological recognition of the sample, which unfortunately can be time-consuming and subjective. Therefore, researchers from CIRI beamline developed a method, IR imaging combined with machine learning, for comprehensive histopathology recognition in pancreatic tissues, and patient classification. The created model allowed for very high accuracy patient classification as cancerous or benign (AUC values above 0.95 – where AUC equal to 1 means perfect classifier). Moreover, they were able to recognize six tissue classes, even in surgical resection biopsy coming from a hospital. They compared also the influence of different spatial resolutions (standard or high definition) on classification results. The main aim of this research was to develop a state-of-the-art baseline model for future clinical applications and was funded by a Homing grant from the Foundation for Polish Science.
Figure 1. Patient pathology recognition pipeline: A) Tissue Micro Array preparation, B) Histopathological annotation on H&E stained biopsies images, C) Biopsies' Infrared Imaging and obtained data structure, D) Model creation and patient pathology recognition.
Written by: Danuta Liberda-Matyja
Link to the publication:
D. Liberda, P. Koziol, T. Wrobel, Comprehensive Histopathology Imaging in Pancreatic Biopsies: High Definition Infrared Imaging with Machine Learning Approach, International Jurnal of Biological Sciences, 2023, 19(10):3200-3208 doi:10.7150/ijbs.83068
