The SOLCRYS beamline at NSRC Solaris will offer routine studies of the protein structure using the diffraction of synchrotron radiation. Currently, the progress in the field of structural biology is based on the coexistence of 3 main research techniques: macromolecular crystallography, cryo-electron microscopy of single molecules and nuclear magnetic resonance spectroscopy. However, macromolecular crystallography is still the main method of determining the spatial structure of proteins and their complexes. Synchrotron beamlines dedicated to protein crystallography are located in almost all synchrotrons in the world. Structural research is carried out on them using serial crystallography techniques or screening of protein complexes with compounds of potential medical importance. Despite the intensive development of cryo-electron microscopy, the study of the structure of protein complexes with nucleic acids or other proteins is still at the center of protein crystallography applications. The structures of macromolecular complexes provide information not only about the molecular architecture but also about interactions between macromolecules.
An example of studies of macromolecular complexes carried out using synchrotron radiation diffraction is the structure of the mitochondrial RNA degradosome complex from Candidia glabrata, consisting of 2 subunits - nuclease and helicase [1]. These enzymes play a key role in the processing and degradation of RNA. Diffraction experiments were carried out at the European Synchrotron Radiation Facility (Grenoble, France) on the ID23-2 beamline.
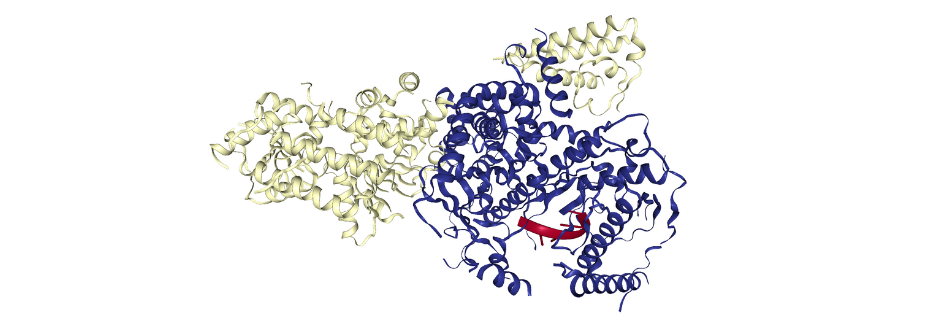
Figure 1. The spatial structure of yeast mitochondrial RNA degradosome complex mtEXO (PDB ID: 6F4A). The mtEXO complex is composed of nuclease (yellow) and helicase (blue) with short RNA fragment (red).
Macromolecular crystallography is often used to characterize the enzymes and their structural homologs. In combination with bioinformatics and other experimental techniques, it is possible to follow the evolution of the proteins in different organisms. Example of such study, performed on the SBC-CAT 19-ID and LS-CAT 21-ID-F beamlines at the Advanced Photon Source (Argonne, USA) presents the crystal structures of two dihydroorotases from Yersinia pestis and Vibrio cholerae [2]. Dihydroorotases are enzymes of de novo pyrimidine biosynthesis pathway that play a key role in the bacterial proliferation. Due to the beamline tunability, it was possible to characterize the metal bound in the active site of the protein (Fig. 2).
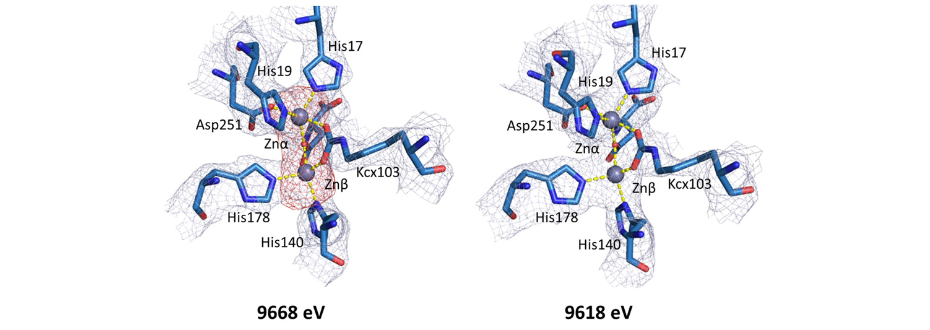
Figure 2. Comparison between electron density maps in active site regions of YpDHO (PDB ID: 6CTY) calculated for datasets collected at energies above (9668 eV) and below the zinc absorption edge (9618 eV). Below the zinc absorption edge, anomalous density map peak (red) corresponding to the position of the zinc metal completely disappears. [2]
[1] M. Razew et al., Structural Analysis of MtEXO Mitochondrial RNA Degradosome Reveals Tight Coupling of Nuclease and Helicase Components, Nat Commun 9, 97 (2018).
[2] J. Lipowska, C. D. Miks, K. Kwon, L. Shuvalova, H. Zheng, K. Lewiński, D. R. Cooper, I. G. Shabalin, and W. Minor, Pyrimidine Biosynthesis in Pathogens – Structures and Analysis of Dihydroorotases from Yersinia Pestis and Vibrio Cholerae, Int J Biol Macromol 136, 1176 (2019).

